06 Jun 2025
Wild bees are indispensable pollinators in ecosystems, but their survival and health are threatened by urbanization-induced habitat fragmentation, pesticide exposure, and resource competition. Recently, Dr. Min Tang's research group, Sustainable2be(e) Lab(https://connect.xjtlu.edu.cn/group/tangmin-biolab), published an innovative study in the well-respected journal Insect Science titled: "Urban wild bee well-being revealed by gut metagenome data: A mason bee model."
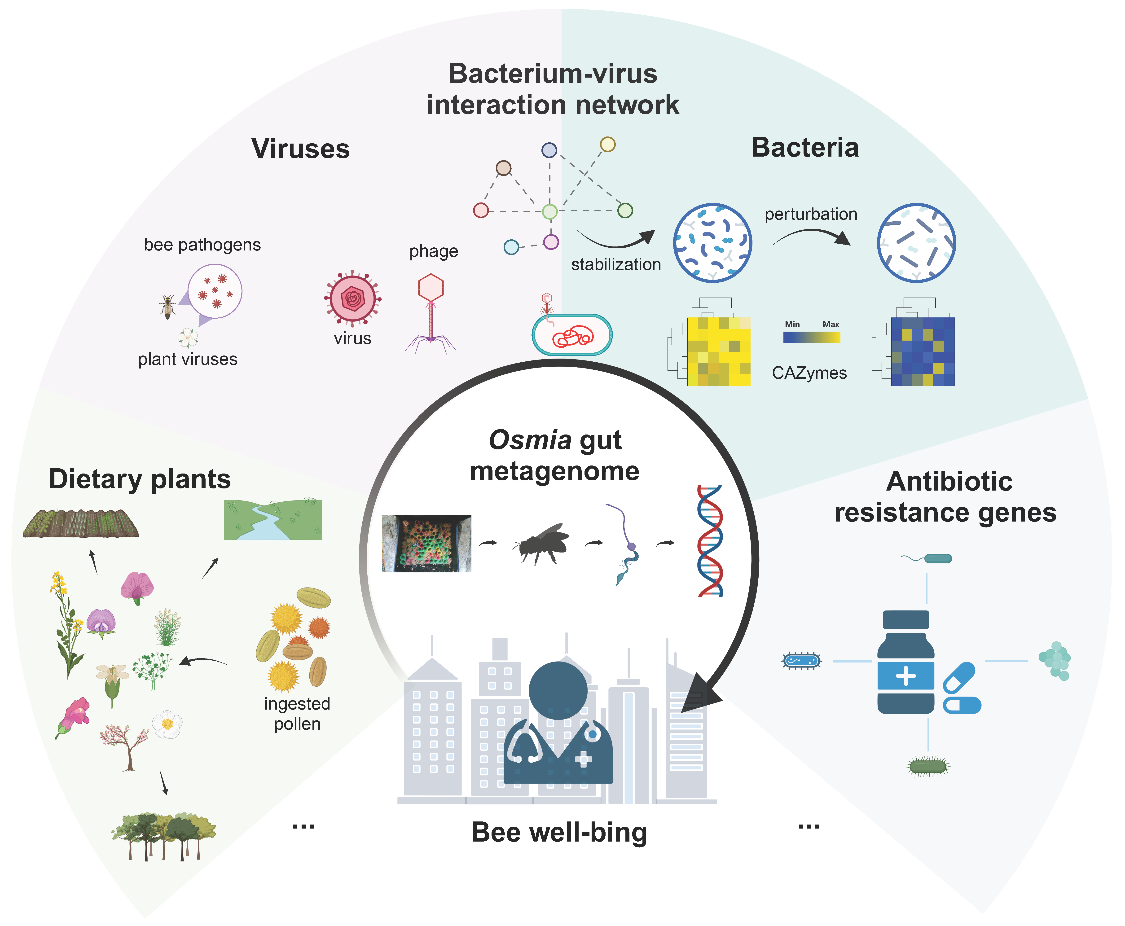
Focusing on a vital native pollinator in China, the mason bee (Osmia excavata), the research employs an integrated metagenomic analysis of diet, bacteriome, resistome, virome, and interaction networks to systematically assess the impact of urban ecological stressors on bee health and microecological homeostasis.
Dr. Min Tang from the Department of Biological Sciences and Bioinformatics at XJTLU's School of Science is the corresponding author, and her Ph.D. student Yiran Li is the first author. Undergraduate students Chengweiran Liu, Yiran Wang, Muhan Li, Xingyu Hu, Zhiwei Chen, and Mingrui Li from the Sustainable2be(e) Lab, as well as Ph.D. students Shasha Zou, Changsheng Ma, and Chinonye Jennifer Obi from the Department of Health and Environmental Sciences, all contributed to this research. Dr. Yi Zou from the Department of Health and Environmental Sciences and Prof. Xin Zhou from China Agricultural University provided significant guidance and support to the study.
Key Findings
Gut Metagenomics Decodes Urban Bee Health
As a typical solitary bee, the mason bee is an important pollinator of fruit crops in China. The research team set up artificial nests at 10 urban agricultural sites in Suzhou to collect samples of Osmia excavata for in-depth analysis.
1.Limited Dietary Diversity in Urban Landscapes
The study found that mason bees in urban areas primarily foraged on crops such as rapeseed (Brassica) and pea (Pisum), as well as urban greening plants like the plane tree (Platanus). Despite providing sustenance, the low floral diversity highlights the need for enhanced nutritional support in urban ecosystems.
2. Gut Microbiome Stability and Perturbations
The core gut bacteriome was dominated by Sodalis and exhibited structural stability. However, at two sites, Sodalis was replaced by non-symbiotic bacteria (e.g., Pseudomonas), accompanied by a significant reduction in carbohydrate-active enzymes (CAZymes)—indicating compromised nutrient metabolism and potential dysbiosis.
3. Environmental Stress Signatures
Antibiotic Resistance Genes (ARGs): 173 ARGs were detected, predominantly multidrug/unclassified types, with site-specific variations suggesting localized antibiotic pollution or microbial selection pressure.
Virome Insights: DNA virome analysis identified abundant bacteriophages (including novel viruses) and honeybee pathogens (Apis mellifera filamentous virus, AmFV), indicating pathogen spillover from managed bees. Sites with dysbiosis lacked temperate phages and showed elevated animal viruses.
4. Network Robustness of Microbiome-
Virome Interactions
Sodalis-centric bacteria-phage networks demonstrated higher stability under simulated ecological disturbances than bacteria-only networks. This underscores the viruses’ role as active regulators of microbial homeostasis, not mere pathogens.
Implications: Gut Metagenomics as an Early-Warning System
This important research study positions wild bee gut metagenomics as a sensitive indicator of urban environmental health. It calls for effective actionable strategies: Designing connected urban green spaces with diverse native flora; Reducing agrochemical/antibiotic usage; and Creating pollinator-friendly habitats to mitigate anthropogenic stressors.
Training Scientific Rigor
From Undergraduate Project to High-Impact Research
This research originated from XJTLU’s 2023 Summer Undergraduate Research Fellowship (SURF) project. Lead author Yiran Li, was a third-year undergraduate student at the time, undertaking her first research task under Dr. Tang’s mentorship. Despite obtaining key results in 2023, Dr. Tang prioritized long-term, high quality scientific training, requiring students to treat every analysis as "publication-ready" through iterative code validation and data verification.

In 2024, undergraduate students Chengweiran Liu and Yiran Wang, joined the team and expanded the study by constructing cross-domain interaction networks. Their analysis of network topology revealed the ecological stability mechanisms of Sodalis-dominated systems, strengthening the research paper’s methodological rigor and ecological narrative.
Reflecting on her journey, now-PhD candidate Yiran Li stated:
"This project transformed my understanding of scientific research: it’s not just knowledge acquisition but continuous application and discovery. Dr. Tang’s rigorous yet open approach, emphasizing independent thinking and long-term growth, deeply resonated with me.”
After a year of intensive training, Yiran declined master’s offers (including one from the Imperial College London) to pursue a PhD at XJTLU, citing enhanced resilience in bioinformatics and critical analysis skills.
The study exemplifies the success of XJTLU’s SURF program in providing significant supports to the scientific growth of undergraduate students by integrating high quality research training with high-impact outputs, driven by Dr. Tang’s philosophy: "Prioritize process, quality, and logical thinking."
Dr. Tang continues to recruit bright, highly motivated, hardworking and dedicated Bachelors, Masters and PhD students to join her growing Research Team and Lab. Those students who are interested are welcome to contact her at Min.Tang@xjtlu.edu.cn.
Original Publication Information:
Li, Y., Liu, C., Wang, Y., et al. (2025). Urban wild bee well-being revealed by gut metagenome data: A mason bee model. Insect Science. DOI: 10.1111/1744-7917.70051
Analysis Code Available at:
https://github.com/Sustainable-to-bee-Lab/wildBeeHealth
Content material: Dr. Min Tang, Yiran Li
Editor and translator: Luyao
Reviewer: Professor John Moraros, Dr. Min Tang
06 Jun 2025








